For the pdf slides, click here
GLM overview
In a GLM, a smooth monotonic link function \(g(\cdot)\) connects the expectation \(\mu_i = E(Y_i)\) with the linear combination of \(\mathbf{X}_i\), \[\begin{equation}\label{eq:glm_link} g(\mu_i) = \eta_i = \mathbf{X}_i \boldsymbol\beta \end{equation}\]
In a generalized linear mixed model (GLMM), we have \[ g(\mu_i) = \eta_i = \mathbf{X}_i \boldsymbol\beta + \mathbf{Z}_i \mathbf{b}, \quad \mathbf{b} \sim \text{N}(\mathbf{0}, \boldsymbol\psi) \]
Theory of GLMs
Exponential family
Exponential family of distributions
- The density function for an exponential family distribution
\[\begin{equation}\label{eq:exponential_family_density}
f(y) = \exp\left\{ \frac{y\theta - b(\theta)}{a(\phi)} + c(y, \phi) \right\}
\end{equation}\]
- \(a, b, c\): arbitrary functions
- \(\phi\): an arbitrary scale parameter
- \(\theta\): the canonical parameter; completely depend on the model parameter \(\boldsymbol\beta\)
Properties about exponential family mean and variance \[\begin{align*} E(Y) & = b^{\prime}(\theta)\\ var(Y) & = b^{\prime\prime}(\theta) a(\phi) \end{align*}\]
- In most practical cases, \(a(\phi) = \phi/\omega\) where \(\omega\) is a known constant
- We define a function \[ V(\mu) = b^{\prime\prime}(\theta) / w, \quad \text{so that } var(Y) = V(\mu) \phi \]
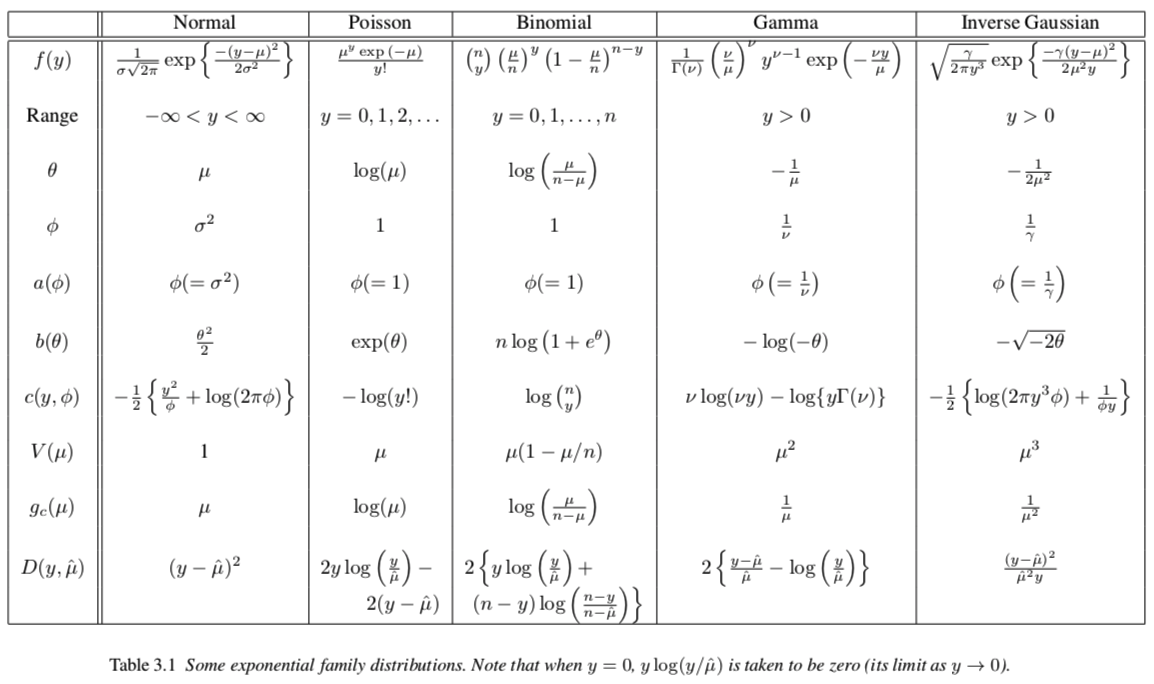
Exponential family examples
Iteratively re-weighted least square (IRLS)
Fitting GLMs
For the GLM model and , assuming \(a_i(\phi) = \phi/\omega_i\), the log likelihood is \[ l(\boldsymbol\beta) = \sum_{i=1}^n \omega_i \left[ y_i \theta_i - b_i(\theta_i) \right]/\phi + c_i(\phi, y_i) \]
- To optimize, we use the Newton’s method, which is an iterative optimization approach
\[
\theta^{(t+1)} = \theta^{(t)} - \left(\nabla^2 l\right)^{-1}\nabla l
\]
- Where both \(\nabla^2 l\) and \(\nabla l\) are evaluated at the current iteration \(\theta^{(t)}\)
- Alternatively, we can use the Fisher scoring variant of the Newton’s method, by replacing the Hessian matrix with its expectation
Next, we will need to compute the gradient vector and expected Hessian matrix of \(l\)
Compute the gradient vector and expected Hessian of \(l\)
By the chain rule, \[\begin{align*} \frac{\partial \theta_i}{\partial \beta_j} & = \frac{\partial \theta_i}{\partial \mu_i} \cdot \frac{\partial \mu_i}{\partial \eta_i} \cdot \frac{\partial \eta_i}{\partial \beta_j} \\ & = \frac{1}{b^{\prime\prime}(\theta_i)}\cdot \frac{1}{g^{\prime}(\mu_i)}\cdot X_{ij} \end{align*}\]
Therefore, the gradient vector of \(l\) is \[\begin{align*} \frac{\partial l}{\partial \beta_j} = \frac{1}{\phi} \sum_{i=1}^n \omega_i \left[y_i - b^{\prime}_i(\theta_i) \right]\frac{\partial \theta_i}{\partial \beta_j} = \frac{1}{\phi}\sum_{i=1}^n \frac{y_i - \mu_i}{g^{\prime}(\mu_i) V(\mu_i)} X_{ij} \end{align*}\]
The expected Hessian (expectation taken wrt \(Y\)) is \[ E\left( \frac{\partial^2 l}{\partial \beta_j \partial \beta_k} \right) = - \frac{1}{\phi}\sum_{i=1}^n \frac{X_{ij} X_{ik}}{g^{\prime}(\mu_i)^2 V(\mu_i)} \]
The Fisher scoring update
Define the matrices \[\begin{align}\label{eq:W} \mathbf{W} & = \text{diag}\{w_i\}, \quad w_i = \frac{1}{g^{\prime}(\mu_i)^2 V(\mu_i)} \\ \label{eq:G} \mathbf{G} & = \text{diag}\left\{g^{\prime}(\mu_i)\right\} \end{align}\]
The Fisher scoring update for \(\boldsymbol\beta\) is \[\begin{align*} \boldsymbol\beta^{(t+1)} & = \boldsymbol\beta^{(t)} + \left(\mathbf{X}^T \mathbf{W} \mathbf{X} \right)^{-1} \mathbf{X}^T \mathbf{W} \mathbf{G} (\mathbf{y} - \boldsymbol\mu)\\ & = \left(\mathbf{X}^T \mathbf{W} \mathbf{X} \right)^{-1} \mathbf{X}^T \mathbf{W} \underbrace{\left[ \mathbf{G}(\mathbf{y} - \boldsymbol\mu) + \mathbf{X}\boldsymbol\beta^{(t)} \right]}_{\mathbf{z}} \end{align*}\]
Iteratively re-weighted least square (IRLS) algorithm
Initialization: \[ \hat{\mu}_i = y_i + \delta_i, \quad \hat{\eta}_i = g\left(\hat{\mu}_i\right) \]
- \(\delta_i\) is usually zero, but may be a small constant ensuring \(\hat{\eta}_i\) is finite
Compute pseudo data \(z_i\) and iterative weights \(w_i\): \[\begin{align*} z_i &= g^{\prime}\left(\hat{\mu}_i\right)\left(y_i - \hat{\mu}_i\right) + \hat{\eta}_i\\ w_i &= \frac{1}{g^{\prime}\left(\hat{\mu}_i\right)^2 V\left(\hat{\mu}_i\right)} \end{align*}\]
Find \(\hat{\boldsymbol\beta}\) by minimizing the weighted least squares objective \[ \sum_{i=1}^n w_i \left(z_i -\mathbf{X}_i \boldsymbol\beta \right)^2 \] then update \[ \hat{\boldsymbol\eta} = \mathbf{X} \hat{\boldsymbol\beta}, \quad \hat{\mu}_i = g^{-1}\left(\hat{\eta}_i\right) \]
- Repeat Step 2-3 until the change in deviance is near zero
IRLS example 1: logistic regression
For logistic regression, \[\begin{align*} g(\mu) & = \log\left(\frac{\mu}{1 - \mu}\right), \quad g^{\prime}(\mu) = \frac{1}{\mu (1-\mu)}\\ V(\mu) &= \mu (1-\mu), \quad\quad\quad \phi =1 \end{align*}\]
Therefore, in Step 2 of IRLS, \[\begin{align*} z_i & = \frac{y_i - \hat{\mu}_i}{\hat{\mu}_i\left(1-\hat{\mu}_i\right)} + \hat{\eta}_i \\ w_i & = \hat{\mu}_i\left(1-\hat{\mu}_i\right) \end{align*}\]
IRLS example 2: GLM with independent normal priors
Assume that the vector \(\boldsymbol\beta\) has independent normal priors \[ \boldsymbol\beta \sim \text{N}\left(\mathbf{0}, \frac{\phi}{\lambda}\mathbf{I}_p \right) \]
Log posterior density (we still call it \(l\), with some abuse of notation) \[ l(\boldsymbol\beta) = \frac{1}{\phi}\sum_{i=1}^n \omega_i \left[ y_i \theta_i - b_i(\theta_i) \right]- \frac{\lambda}{2\phi}\boldsymbol\beta^T \boldsymbol\beta + \text{const} \]
Gradient vector and expected Hessian matrix (wrt \(\boldsymbol\beta\)) \[\begin{align*} \nabla l &= \frac{1}{\phi} \left[\mathbf{X}^T \mathbf{W}\mathbf{G}(\mathbf{y} - \boldsymbol\mu) - \lambda \boldsymbol\beta\right]\\ E\left(\nabla^2 l \right) &= -\frac{1}{\phi}\left(\mathbf{X}^T \mathbf{W}\mathbf{X} + \lambda \mathbf{I}_p \right) \end{align*}\]
- Here, \(\mathbf{W}\) and \(\mathbf{G}\) are the same as in Equation and
- IRLS for GLM with independent normal priors \[\begin{align}\notag \boldsymbol\beta^{(t+1)} & = \boldsymbol\beta^{(t)} + \left(\mathbf{X}^T \mathbf{W} \mathbf{X} + \lambda \mathbf{I}_p \right)^{-1} \left[\mathbf{X}^T \mathbf{W} \mathbf{G}(\mathbf{y} - \boldsymbol\mu) - \lambda \boldsymbol\beta^{(t)}\right] \\ \label{eq:IRLS_penalty} & = \left(\mathbf{X}^T \mathbf{W} \mathbf{X} + \lambda \mathbf{I}_p\right)^{-1} \mathbf{X}^T \mathbf{W} \underbrace{\left[ \mathbf{G}(\mathbf{y} - \boldsymbol\mu) + \mathbf{X}\boldsymbol\beta^{(t)} \right]}_{\mathbf{z}} \end{align}\]
Asymptotic consistency of MLE, deviance, tests, residuals
Large sample distribution of \(\hat{\boldsymbol\beta}\)
Hessian of the negative log likelihood (also called observed information) \[ \hat{\mathcal{I}} = \frac{\mathbf{X}^T \mathbf{W} \mathbf{X}}{\phi} \]
Fisher information, also called expected information \[ \mathcal{I} = E\left(\hat{\mathcal{I}}\right) \]
Asymptotic normality the MLE \(\hat{\boldsymbol\beta}\) \[ \hat{\boldsymbol\beta} \sim \text{N}\left(\boldsymbol\beta, \mathcal{I}^{-1} \right) \quad \text{or} \quad \hat{\boldsymbol\beta} \sim \text{N}\left(\boldsymbol\beta, \hat{\mathcal{I}}^{-1} \right) \]
Deviance
Deviance is the GLM counterpart of the residual sum of squares in normal linear regression \[\begin{align}\notag D & = 2\phi \left[ l\left(\hat{\boldsymbol\beta}_{\max} \right) - l\left(\hat{\boldsymbol\beta} \right) \right]\\ \label{eq:deviance} &= \sum_{i=1}^n 2 \omega_i \left[y_i \left(\tilde{\theta}_i - \hat{\theta}_i \right) - b\left(\tilde{\theta}_i \right) + b\left(\hat{\theta}_i \right) \right] \end{align}\]
- Here, \(l\left(\hat{\boldsymbol\beta}_{\max} \right)\) is the maximized likelihood of the saturated model: the model with one parameter per data point. For exponential family distribution, it is computed by simply setting \(\hat{\boldsymbol\mu} = \mathbf{y}\).
- \(\tilde{\boldsymbol\theta}\) and \(\hat{\boldsymbol\theta}\) are the maximum likelihood estimates of the canonical parameters for the saturated model and the model of interest, respectively
From the second equality, we can see that deviance is independent of \(\phi\)
For normal linear regression, deviance equals the residual sum of squares
Scaled deviance
Scaled deviance does depend on \(\phi\) \[ D^* = \frac{D}{\phi} \]
If the model is specified correctly, then approximately \[ D^* \sim \chi^2_{n-p} \]
- To compare two nested models,
- If \(\phi\) is known, then under \(H_0\), we can use \[ D^*_0 - D^*_1 \sim \chi^2_{p_1 - p_0} \]
- If \(\phi\) is unknown, then under \(H_0\), we can use \[ F = \frac{(D_0 - D_1)/(p_1 - p_0)}{D_1/(n - p_1)} \sim F_{p_1 - p_0, n - p_1} \]
Canonical link functions
The canonical link \(g_c\) is the link function such that \[ g_c(\mu_i) = \theta_i = \eta_i \] where \(\theta_i\) is the canonical parameter of the distribution
Under canonical links, the observed information \(\hat{\mathcal{I}}\) and the expected information \(\mathcal{I}\) matrices are the same
Under canonical links, since \(\frac{\partial \theta_i}{\partial \beta_j} = X_{ij}\), the system of equations that the MLE satisfies becomes \[ \frac{\partial l}{\partial \beta_j} = \sum_{i=1}^n \omega_i (y_i - \mu_i) X_{ij} = 0 \] Thus, if \(\omega_i =1\), we have \[ \mathbf{X}^T \mathbf{y} = \mathbf{X}^T \hat{\boldsymbol\mu} \]
- For any GLM with an intercept term and canonical link, the residuals sum to zero, i.e., \(\sum_i y_i = \sum_i \hat{\mu}_i\)
GLM residuals
Model checking is perhaps the most important part of applied statistical modeling
It is usual to standardize GLM residuals so that if the model assumptions are correct,
- the standardized residuals should have approximately equal variance, and
- behave like residuals from an ordinary linear model
Pearson residuals \[ \hat{\epsilon}_i^p = \frac{y_i - \hat{\mu}_i} {\sqrt{V\left(\mu_i\right)}} \]
- In practice, the distribution of the Pearson residuals can be quite asymmetric around zero. So the deviance residuals (introduced next) are often preferred.
Deviance residuals
Denote \(d_i\) as the \(i\)th component in the deviance definition , so that the deviance is \(D = \sum_{i=1}^n d_i\)
By analogy with the ordinary linear model,we define the deviance residual \[ \hat{\epsilon}_i^d = \text{sign}(y_i - \hat{\mu_i}) \sqrt{d_i} \]
- The sum of squares of the deviance residuals gives the deviance itself
Quasi-likelihood (GEE)
Quasi-likelihood
Consider an observation \(y_i\), of a random variable with mean \(\mu_i\) and known variance function \(V(\mu_i)\)
- Getting the distribution of \(Y_i\) exactly right is rather unimportant, as long as the mean-variance relationship \(V(\cdot)\) is correct
Then the log quasi-likelihood for \(\mu_i\), given \(y_i\), is \[ q_i(\mu_i) = \int_{y_i}^{\mu_i} \frac{y_i - z}{\phi V(z)}~dz \]
- The log quasi-likelihood for the mean vector \(\boldsymbol\mu\) of all the response data is \(q(\boldsymbol\mu) = \sum_{i=1}^n q_i (\mu_i)\)
To obtain the maximum quasi-likelihood estimation of \(\boldsymbol\beta\), we can differentiate \(q\) wrt \(\beta_j\), for \(\forall j\) \[ 0 = \frac{\partial q}{\partial \beta_j} = \sum_{i=1}^n \frac{y_i - \mu_i}{\phi V(\mu_i)} \frac{\partial \mu_i}{\partial \beta_j} \Longrightarrow \sum_{i=1}^n \frac{y_i - \mu_i}{V(\mu_i) g^{\prime}(\mu_i)} X_{ij} = 0 \] this is exactly the GLM maximum likelihood solution, which can be obtained through IRLS
Generalized Linear Mixed Models (GLMM)
Generalized linear mixed models (GLMM)
A GLMM model for an exponential family random variable \(Y_i\) \[ g(\mu_i) = \mathbf{X}_i \boldsymbol\beta + \mathbf{Z}_i \mathbf{b}, \quad \mathbf{b} \sim \text{N}\left(\mathbf{0}, \boldsymbol\psi_{\boldsymbol\theta} \right) \]
Difficulty in moving from linear mixed models to GLMM: it is no longer possible to evaluate the marginal likelihood analytically
One effective solution is Taylor expansion around \(\hat{\mathbf{b}}\), the posterior mode of \(f(\mathbf{b}\mid \mathbf{y}, \boldsymbol\beta)\)
\[\begin{align*} f(\mathbf{y} \mid \boldsymbol\beta)\approx &\int \exp\left\{ \log f(\mathbf{y}, \hat{\mathbf{b}} \mid \boldsymbol\beta)\right.\\ &~~~~~~~~~~~~ + \left. \frac{1}{2}\left(\mathbf{b} - \hat{\mathbf{b}}\right)^T\frac{\partial^2 \log f(\mathbf{y}, \mathbf{b}\mid \boldsymbol\beta)}{\partial\mathbf{b}\partial \mathbf{b}^T}\left(\mathbf{b} - \hat{\mathbf{b}}\right)\right\} d\mathbf{b} \end{align*}\]
Laplace approximation of GLMM marginal likelihood
For GLM, note that the expected Hessian is \[ -\frac{\mathbf{Z}^T \mathbf{W}\mathbf{Z}}{\phi} - \boldsymbol\psi^{-1} \]
- \(\mathbf{W}\) is the IRLS weight vector based on the \(\boldsymbol\mu\) implied by \(\hat{\mathbf{b}}\) and \(\boldsymbol\beta\)
Therefore, the approximate marginal likelihood is \[ f(\mathbf{y} \mid \boldsymbol\beta)\approx f(\mathbf{y}, \hat{\mathbf{b}} \mid \boldsymbol\beta)\frac{(2\pi)^{p/2}}{\left| \frac{\mathbf{Z}^T \mathbf{W}\mathbf{Z}}{\phi} + \boldsymbol\psi^{-1}_{\boldsymbol\theta} \right|^{1/2}} \]
Penalized likelihood and penalized IRLS
- The point estimators \(\hat{\boldsymbol\beta}\) and \(\hat{\mathbf{b}}\) are obtained by optimizing the penalized likelihood
\[\begin{align*} \hat{\boldsymbol\beta}, \hat{\mathbf{b}} &= \arg\max_{\boldsymbol\beta, \mathbf{b}} \log f(\mathbf{y}, \mathbf{b} \mid \boldsymbol\beta)\\ &= \arg\max_{\boldsymbol\beta, \mathbf{b}} \left\{\log f(\mathbf{y} \mid \mathbf{b}, \boldsymbol\beta) - \mathbf{b}^T \boldsymbol\psi^{-1}_{\mathbf{\theta}}\mathbf{b}/2\right\} \end{align*}\]
To simplify notation, we denote \[\begin{align*} \mathcal{B}^T & = (\mathbf{b}, \boldsymbol\beta)^T\\ \mathcal{X} & = (\mathbf{Z}, \mathbf{X}), \quad \mathbf{S} = \left[ \begin{array}{cc} \boldsymbol\psi^{-1}_{\mathbf{\theta}} & \mathbf{0}\\ \mathbf{0} & \mathbf{0} \end{array} \right] \end{align*}\]
A penalized version of the IRLS algorithm (PIRLS) : by , a single Newton update step is \[\begin{align*} \mathcal{B}^{(t+1)} = \left( \mathcal{X}^T \mathbf{W} \mathcal{X} + \phi \mathbf{S}\right)^{-1} \mathcal{X}^T \mathbf{W} \left[\mathbf{G}\left(\mathbf{y} - \hat{\boldsymbol\mu}\right)+ \mathcal{X}\mathcal{B}^{(t)} \right] \end{align*}\]
Penalized quasi-likelihood method
- Since optimizing the Laplace approximate marginal likelihood can be computationally costly, it is therefore tempting to instead perform a PIRLS iteration, estimating \(\boldsymbol\theta, \phi\) at each step based on the working mixed model \[ \mathbf{z} \mid \mathbf{b}, \boldsymbol\beta \sim \text{N}\left(\mathbf{X}\boldsymbol\beta + \mathbf{Z}\mathbf{b}, \mathbf{W}^{-1}\phi \right), \quad \mathbf{b} \sim \text{N}\left(\mathbf{0}, \boldsymbol\psi_{\boldsymbol\theta}\right) \]
References
- Wood, Simon N. (2017), Generalized Additive Models: An Introduction with R. Chapman and Hall/CRC