For the pdf slides, click here
Classical (Before Computer Age) Multiple Testing Corrections
Background and notations
Before computer age, multiple testing may only involve 10 or 20 tests. With the emerge of biomedical (microarray) data, multiple testing may need to evaluate several thousands of tests
Notations
- : total number of tests, e.g., number of genes.
- : the z-statistic of the -th test. Note that if we perform tests other than z-test, say a t-test, then we can use inverse-cdf method to transform the t-statistic into a z-statistic, like below where is the standard normal cdf, and is a t distribution cdf.
- : the indices of the true , having members. Usually, majority of hypotheses are null, so is close to 1.
Hypotheses: standard normal vs normal with a non-zero mean where is the effect size for test
Example: the prostate data
A microarray data of
- people, 52 prostate cancer patients and 50 normal controls
- genes
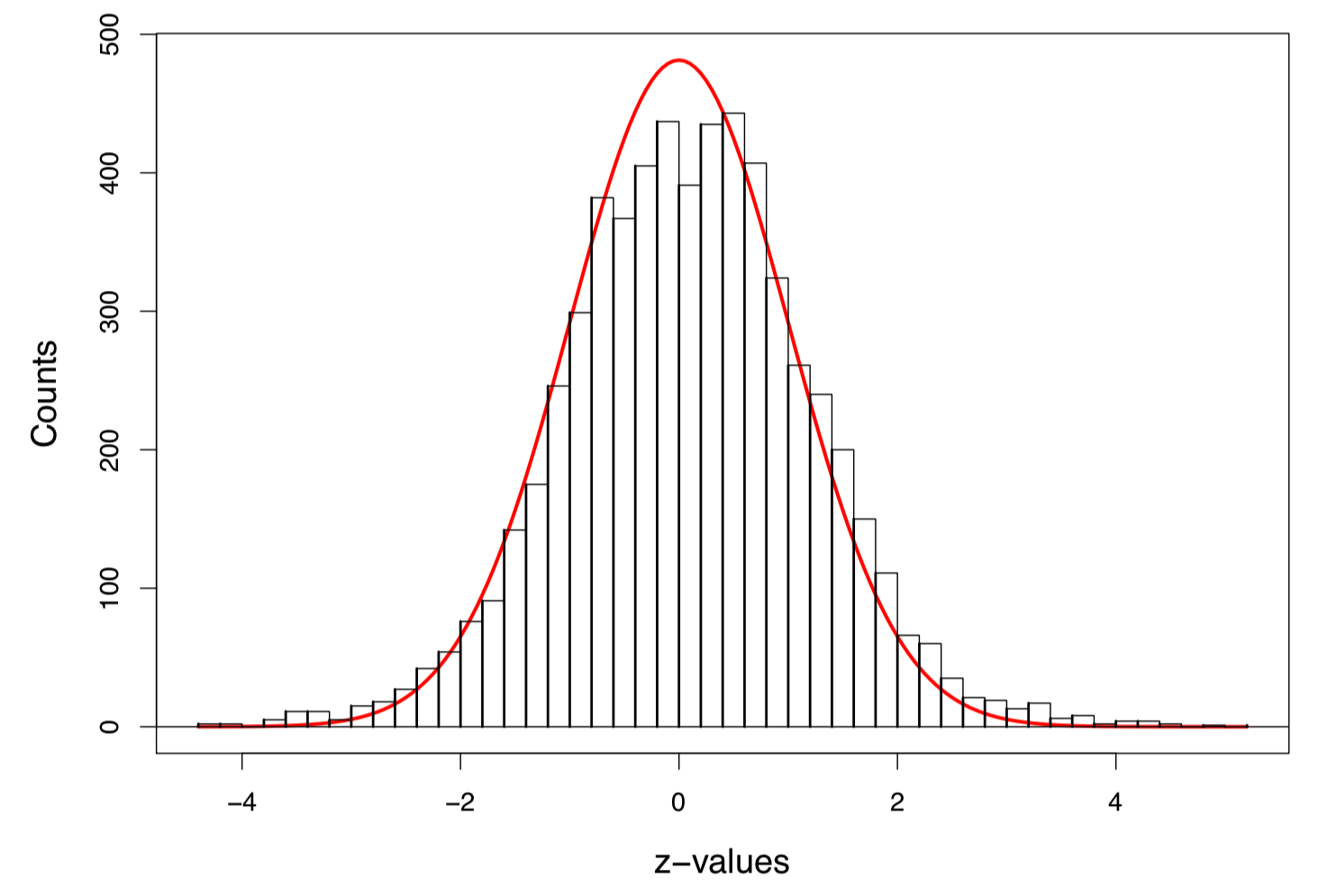
Figure 1: Histogram of 6033 z-values, with the scaled standard normal density curve in red
Bonferroni Correction
Classical multiple testing method 1: Bonferroni bound
- For an overall significance level (usually ), with simultaneous tests, the Bonferroni bound rejects the th null hypothesis at individual significance level
Bonferroni bound is quite conservative!
- For prostate data and , the -value rejection cutoff is very small:
Family-wise Error Rate
Classical multiple testing method 2: FWER control
The family-wise error rate is the probability of making even one false rejection
Bonferroni’s procedure controls FWER, i.e., Bonferroni bound is more conservative than FWER control
FWER control: Holm’s procedure
Order the observed -values from smallest to largest
Reject null hypotheses if
- FWER is usually still too conservative for large , since it was originally developed for
An R function to implement Holm’s procedure
## A function to obtain Holm's procedure p-value cutoff
holm = function(pi, alpha=0.1){
N = length(pi)
idx = order(pi)
reject = which(pi[idx] <= alpha/(N - 1:N + 1))
return(idx[reject])
}## Download prostate data's z-values
link = 'https://web.stanford.edu/~hastie/CASI_files/DATA/prostz.txt'
prostz = c(read.table(link))$V1
## Convert to p-values
prostp = 1 - pnorm(prostz)Illustrate Holm’s procedure on the prostate data
## Apply Holm's procedure on the prostate data
results = holm(prostp)
## Total number of rejected null hypotheses
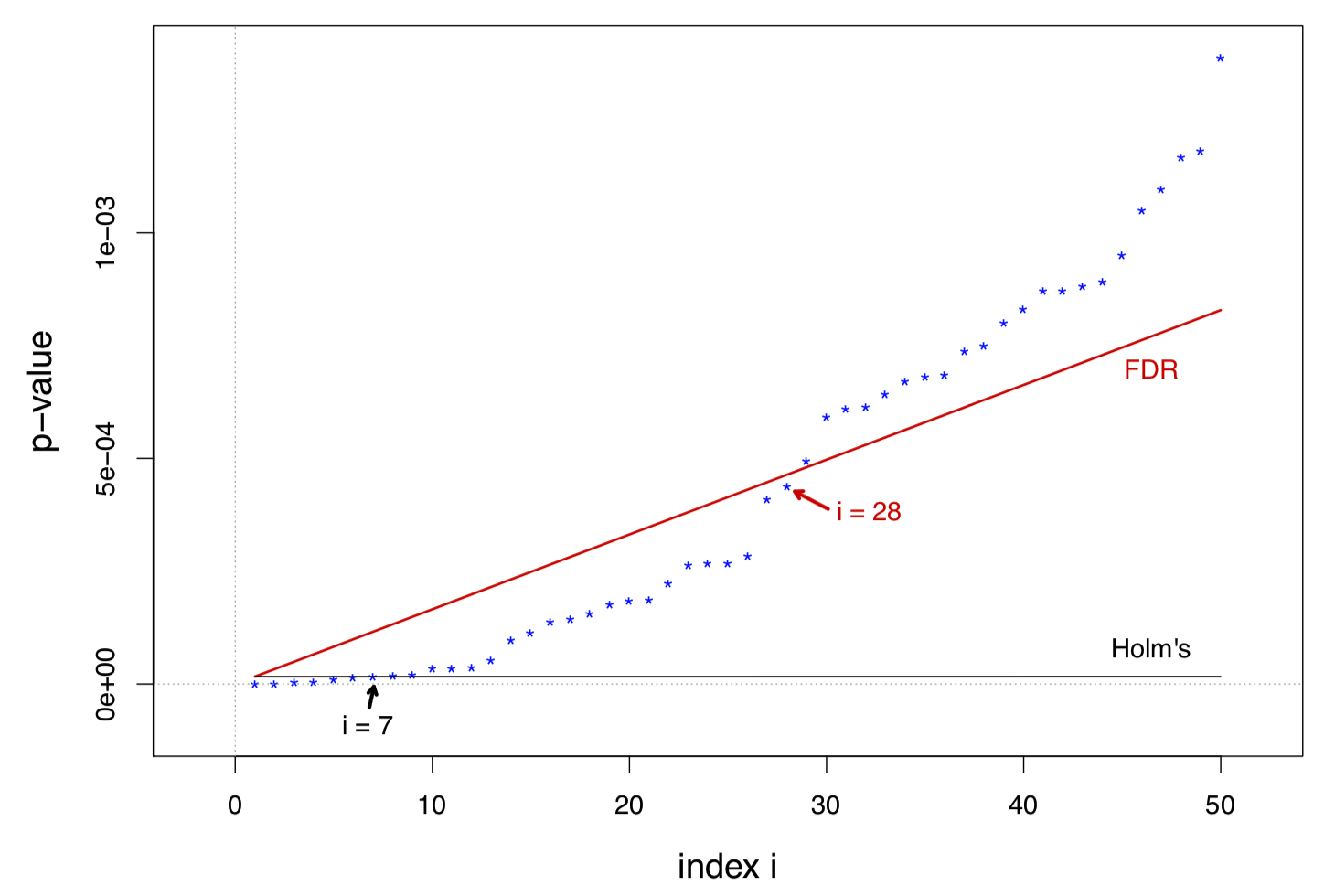
r = length(results); r## [1] 6## The largest z-value among non-rejected nulls
sort(prostz, decreasing = TRUE)[r + 1]## [1] 4.13538## The smallest p-value among non-rejected nulls
sort(prostp)[r + 1]## [1] 1.771839e-05False Discovery Rates
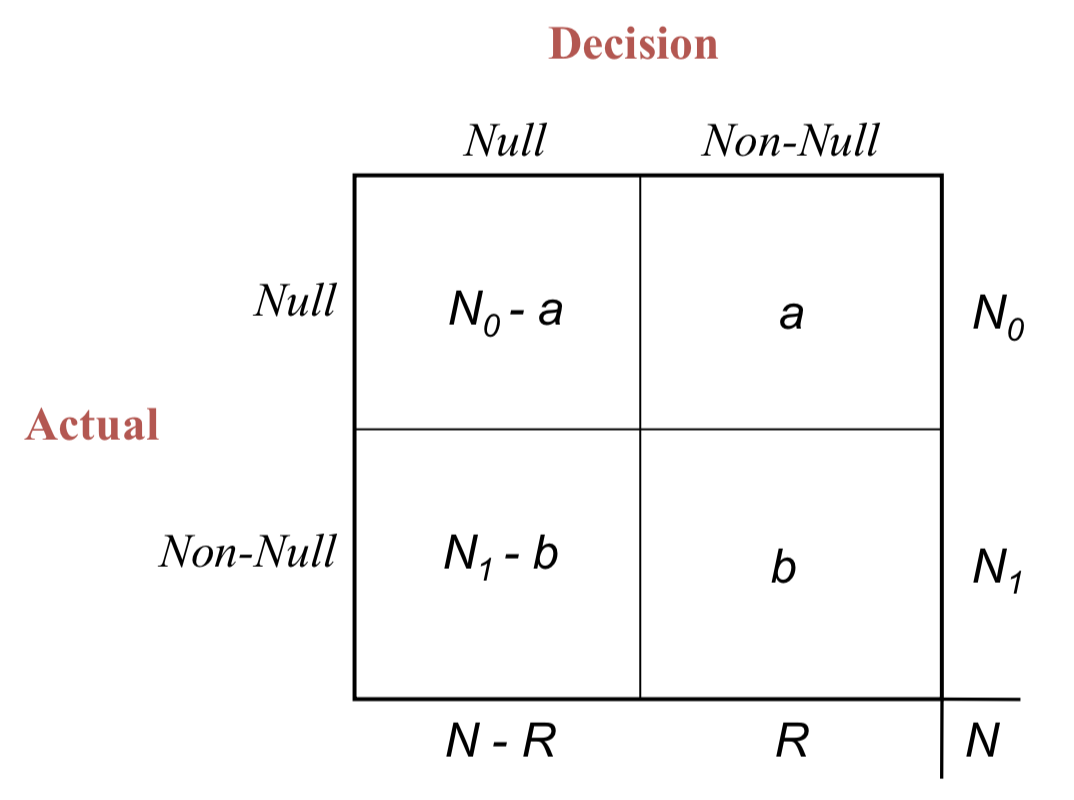
False discovery proportion
FDR control is a more liberal criterion (compared with FWER), thus it has become standard for large multiple testing problems.
- False discovery proportion
- A decision rule rejects out of null hypotheses
- of those are false discoveries (unobservable)
False discovery rate
False discovery rates
A decision rule controls FDR at level , if
- is a prechosen value between 0 and 1
Benjamini-Hochberg FDR control
Benjamini-Hochberg FDR control
Order the observed -values from smallest to largest
Reject null hypotheses if
Default choice
Theorem: if the -values are independent of each other, then the above procedure controls FDR at level , i.e.,
- Usually, majority of the hypotheses are truly null, so is near 1
An R function to implement Benjamini-Hochberg FDR control
## A function to obtain Holm's procedure p-value cutoff
bh = function(pi, q=0.1){
N = length(pi)
idx = order(pi)
reject = which(pi[idx] <= q/N * (1:N))
return(idx[reject])
}Illustrate Benjamini-Hochberg FDR control on the prostate data
## Apply Holm's procedure on the prostate data
results = bh(prostp)
## Total number of rejected null hypotheses
r = length(results); r## [1] 28## The largest z-value among non-rejected nulls
sort(prostz, decreasing = TRUE)[r + 1]## [1] 3.293507## The smallest p-value among non-rejected nulls
sort(prostp)[r + 1]## [1] 0.0004947302Comparing Holm’s FWER control and Benjamini-Hochberg FDR control
In the usual range of interest, large and small , the ratio increases with almost linearly
The figure below is about the prostate data, with
Question about the FDR control procedure
Is controlling a rate (i.e., FDR) as meaningful as controlling a probability (of Type 1 error)?
How should be chosen?
The control theorem depends on independence among the -values. What if they’re dependent, which is usually the case?
The FDR significance for one gene depends on the results of all other genes. Does this make sense?
An empirical Bayes view
Two-groups model
Each of the cases (e.g., genes) is
- either null with prior probability ,
- or non-null with probability
For case , its -value under for has density , cdf , and survival curve
The mixture survival curve
Bayesian false-discovery rate
Suppose the observation for case is seen to exceed some threshold value (say ). By Bayes’ rule, the Bayesian false-discovery rate is
The “empirical” Bayes reflects in the estimation of the denominator: when is large,
An empirical Bayes estimate of the Bayesian false-discovery rate
Connection between and FDR controls
Since and , the FDR control algorithm becomes After rearranging the above formula, we have its Bayesian Fdr bounded
The FDR control algorithm is in fact rejecting those cases for which the empirical Bayes posterior probability of nullness is too small
Answer the 4 questions about the FDR control
(Rate vs probability) FDR control does relate to the posterior probability of nullness
(Choice of ) We can set according to the maximum tolerable amount of Bayes risk of nullness, usually after taking in
(Independence) Most often the , and hence the , are correlated. However even under correlation, is still an unbiased estimator for , making nearly unbiased for .
- There is a price to be paid for correlation, which increases the variance of and
(Rejecting one test depending on others) In the Bayes two-group model, the number of null cases exceeding some threshold has fixed expectation . So an increase in the number of exceeding must come from a heavier right tail for , implying a greater posterior probability of non-nullness .
- This emphasizes the “learning from the experience of others” aspect of empirical Bayes inference
Local False Discovery Rates
Local false discovery rates
Having observed test statistic equal to some value , we should be more interested in the probability of nullness given than
Local false discovery rate
After drawing a smooth curve through the histogram of the -values, we get the estimate
- the null proportion can either be estimated or set equal to 1
A fourth-degree log polynomial Poisson regression fit to the histogram, on the prostate data
Solid line is the local and dashed lines are tail-area
27 genes on the right and 25 one the left have
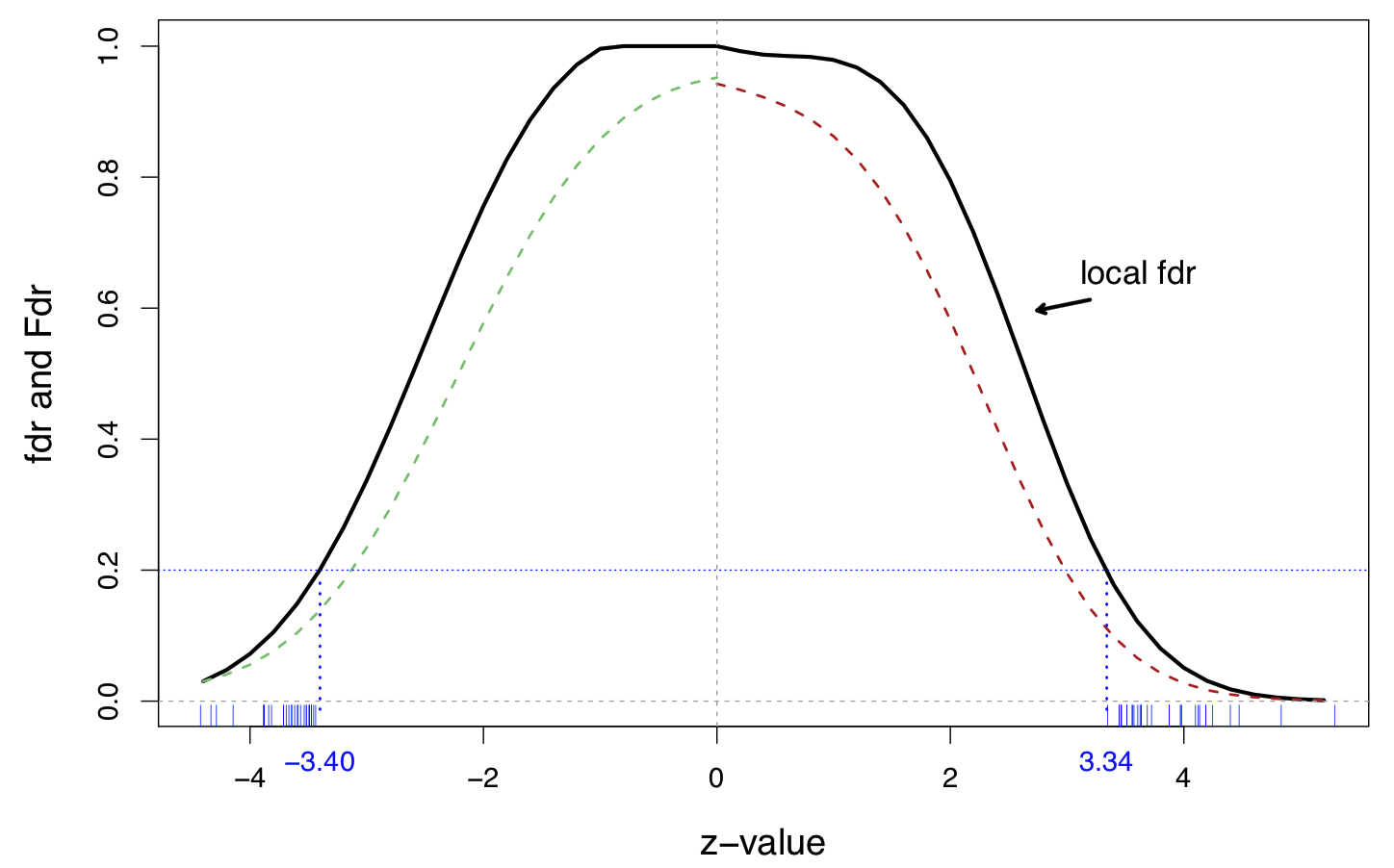
The default cutoff for local fdr
The cutoff is equivalent to
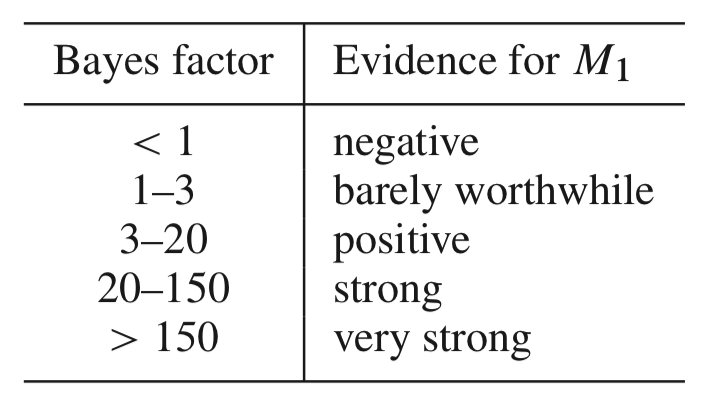
Assuming , this makes the factor factor quite large This is “strong evidence” against the null hypothesis in Jeffrey’s scale of evidence for the interpretation of Bayes factors
Relation between the local and tail-area fdr’s
Since Therefore
Thus, the conventional significant cutoffs are
Empirical Null
Empirical null
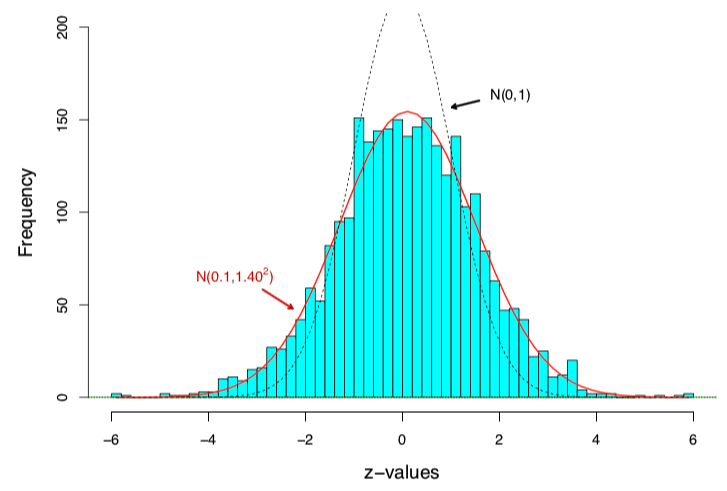
Large scale applications may allow us to empirically determine a more realistic null distribution than
In the police data, a curve is too narrow for the null. Actually, an MLE fit to central data gives as the empirical null
Empirical null estimation
The theoretical null is not completely wrong, but needs adjustment for the dataset at hand
Under the two-group model, with normal but not necessarily standard normal to compute the local , we need to estimate three parameters
Our key assumption is that is large, say , and most of the near are null.
The algorithm
locfdrbegins by selecting a set near and assumes that all the in are nullMaximum likelihood based on the numbers and values of in yield the empirical null estimates
References
Efron, Bradley and Hastie, Trevor (2016), Computer Age Statistical Inference. Cambridge University Press
Links to the prostate data
- The data matrix: prostmat.csv
- The z-values: prostz.txt
A list of FDR methods in R: http://www.strimmerlab.org/notes/fdr.html