For the pdf slides, click here
Introduction of GAM
In general the GAM model has a following structure
- follows some exponential family distribution:
- is a row of the model matrix, and is the corresponding parameter vector
- are smooth functions of the covariates
- This chapter
- Illustrates GAMs by basis expansions, each with a penalty controlling function smoothness
- Estimates GAMs by penalized regression methods
Takeaway: technically GAMs are simply GLM estimated subject to smoothing penalties
Univariate Smoothing
Piecewise linear basis: tent functions
Representing a function with basis expansions
Let’s consider a model containing one function of one covariate
If is the th basis function, then is assumed to have a representation with some unknown parameters
- This is clearly a linear model
The problem with polynomials
A th order polynomial is
The polynomial oscillates wildly in places, in order to both interpolate the data and to have all derivatives wrt continuous
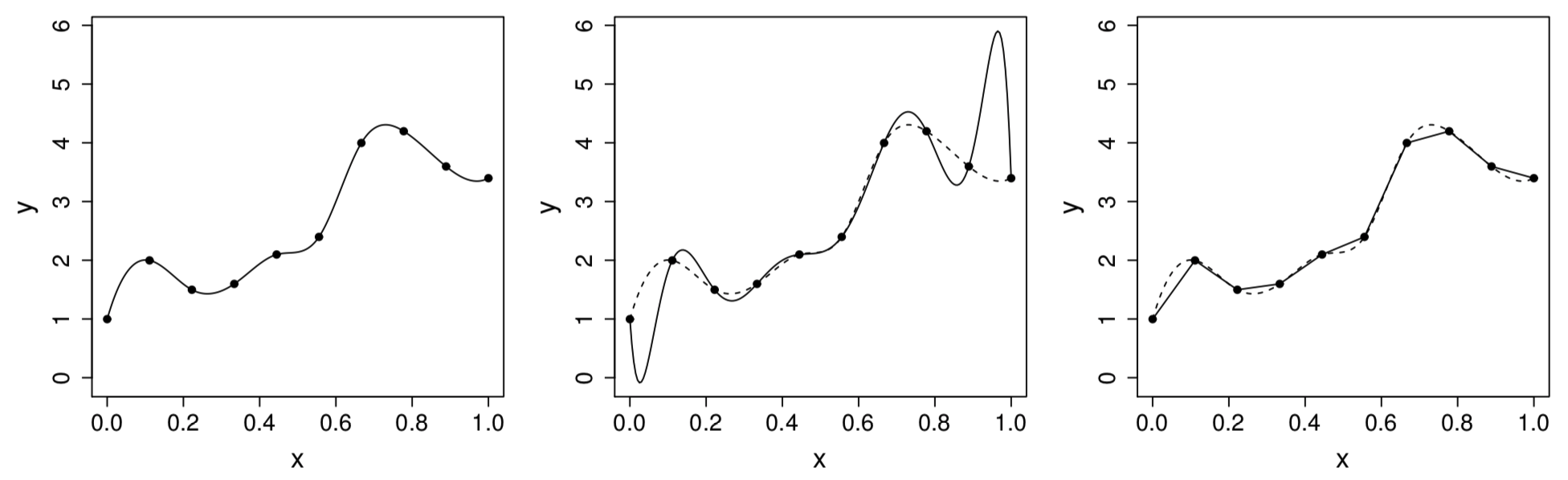
Figure 1: Left: the target function . Middle: polynomial interpolation. Right: piecewise linear interpolant
Piecewise linear basis
Suppose there are knots
The tent function representation of piecewise linear basis is
- For ,
- For the two basis functions on the edge
Visualization of tent function basis
is zero everywhere, except over the interval between the knots immediately to either side of
increases linear from at to 1 at , and then decreases linearly to at
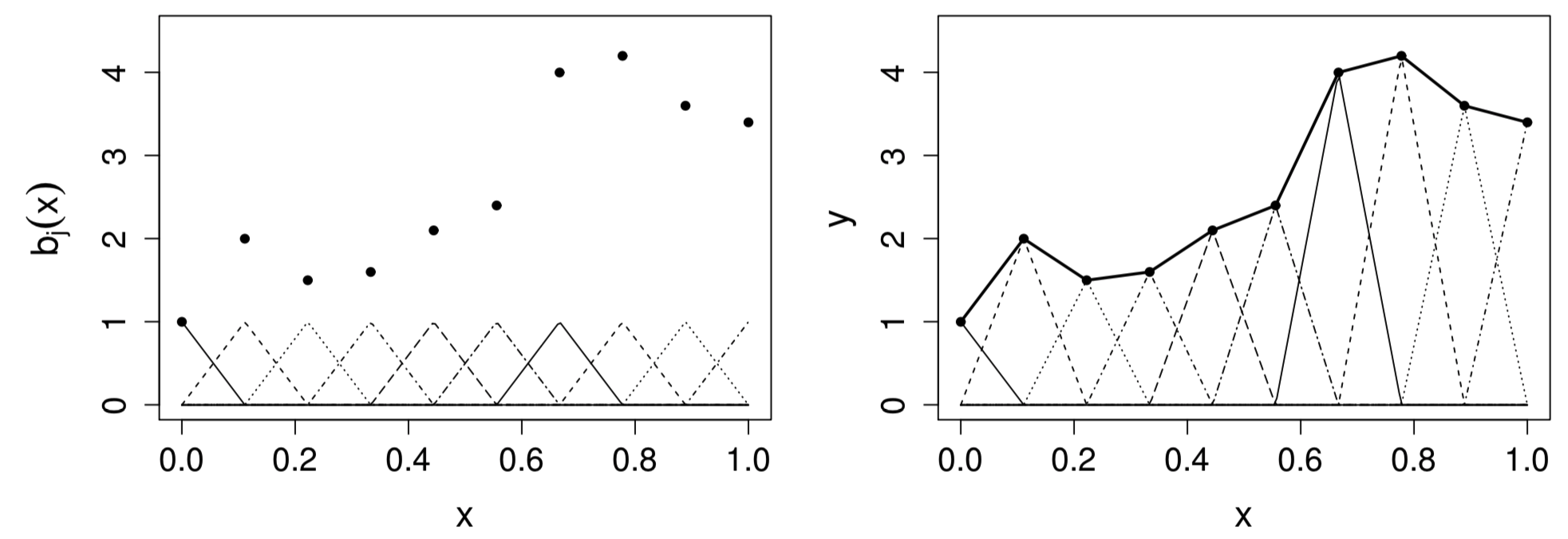
Figure 2: Left: tent function basis, for interpolating the data shown as black dots. Right: the basis functiosn are each multiplied by a coefficient, before being summed
Penalty to control wiggliness
Control smoothness by penalizing wiggliness
To choose the degree of smoothness, rather than selecting the number of knots , we can use a relatively large , but control the model’s smoothness by adding a “wiggliness” penalty
- Note that a model based on evenly spaced knots will not be nested within a model based on evenly spaced knots
Penalized likelihood function for piecewise linear basis:
- Wiggliness is measured as a sum of squared second differences of the function at the knots
- This crudely approximates the integrated squared second derivative penalty used in cubic spline smoothing
- is called the smoothing parameter
Simplify the penalized likelihood
For the tent function basis,
Therefore, the penalty can be expressed as a quadratic form
- The matrix is
- is a square matrix
Solution of the penalized regression
To minimize the penalized likelihood
The hat matrix (also called influence matrix) is thus and the fitted expectation is
For practical computation, we can introduce imaginary data to re-formulate the penalized least square problem to be a regular least square problem
Hyper-parameter tuning
Between the two hyper-parameters: number of knots and the smoothing parameter , the choice of plays the crucial role
We can always use a large enough, more flexible then we expect to need to represent
In
mgcvpackage, the default choice is , and knots are evenly spread out over the range of observed data
Choose by leave-one-out cross validation
Under linear regression, to compute leave-one-out cross validation error (called the ordinary cross validation score), we only need to fit the full model once
- is the model fitted to all data except
- is the model fitted to all data, and is the th diagonal entry of the corresponding hat matrix
In practice, are often replaced by their mean . This results in the generalized cross validation score (GCV)
From the Bayesian perspective
The wiggliness penalty can be viewed as a normal prior distribution on
- Because is rank deficient, the prior covariance is proportional to the pseudo-inverse
The posterior of is still normal
Given the model this extra structure opens up the possibility of estimating and using marginal likelihood maximization or REML (aka empirical Bayes)
Additive Models
A simple additive model with two univariate functions
Let’s consider a simple additive model
The assumption of additive effects is a fairly strong one
The model now has an identifiability problem: and are each only estimable to within an additive constant
- Due to the identifiability issue, we need to use penalized regression splines
Piecewise linear regression representation
Basis representation of and
- The basis functions and are tent functions, with evenly spaced knots and , respectively
Matrix representations
Sum-to-zero constrains to resolve identifiability issues
We assume This is equivalent to for all , which implies
To achieve this condition, we can center the column of
Column centering reduces the rank of to , so that only elements of the vector can be uniquely estimated
- A simple identifiability constraint:
- Set a single element of to zero
- And delete the corresponding column of and
For notation simplicity, in what follows the tildes will be dropped, and we assume that the , are the constrained versions
Penalized piecewise regression additive model
We rewrite the penalized regression as where and
Wiggliness penalties
Fitting additive models by penalized least squares
Penalized least squares objective function
Coefficient estimator
Hat matrix
Conditional posterior distribution
Choosing two smoothing parameters
Since we now have two smoothing parameters , grid searching for the GCV optimal values starts to become inefficient
Instead, R function
optimcan be used to minimize the GCV scoreWe can use log smoothing parameters for optimization to ensure that estimated smoothing parameters are non-negative
Generalized Additive Models
Generalized additive models
Generalized additive models (GAMs): additive models GLM
Penalized iterative least squares (PIRLS) algorithm: iterate the following steps to convergence
Given the current and , compute
Let , we obtain the new by minimizing
Introducing Package mgcv
Introducing package mgcv
Main function:
gam(), very much like theglm()functionSmooth terms:
s()for univariate functions andte()for tensorsA gamma regression example
library(mgcv) ## load the package data(trees)
ct1 <- gam(Volume ~ s(Height) + s(Girth),
family=Gamma(link=log),data=trees)- By default, the degree of smoothness of the is estimated by GCV
summary(ct1)##
## Family: Gamma
## Link function: log
##
## Formula:
## Volume ~ s(Height) + s(Girth)
##
## Parametric coefficients:
## Estimate Std. Error t value Pr(>|t|)
## (Intercept) 3.27570 0.01492 219.6 <2e-16 ***
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## Approximate significance of smooth terms:
## edf Ref.df F p-value
## s(Height) 1.000 1.000 31.32 7.07e-06 ***
## s(Girth) 2.422 3.044 219.28 < 2e-16 ***
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## R-sq.(adj) = 0.973 Deviance explained = 97.8%
## GCV = 0.0080824 Scale est. = 0.006899 n = 31Parital residuals plots
- Pearson residuals added to the estimated smooth terms
par(mfrow = c(1, 2))
plot(ct1,residuals=TRUE)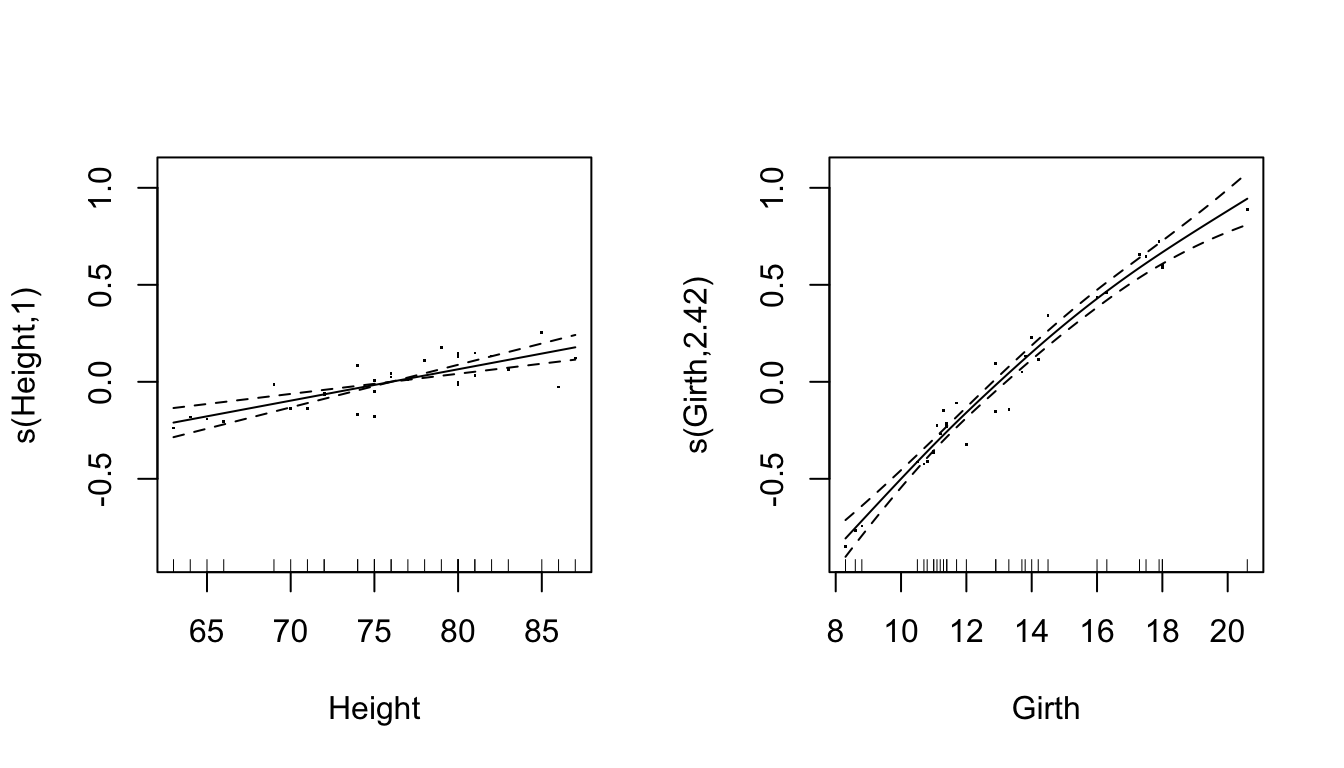 * The number in the -axis label: effective degrees of freedom
* The number in the -axis label: effective degrees of freedom
Finer control of gam(): choice of basis functions
Default: thin plat regression splines
- It has some appealing properties, but can be somewhat computationally costly for large dataset
We can select penalized cubic regression spline by using
s(..., bs = "cr")We can change the dimension of the basis
- The actual effective degrees of freedom for each term is usually estimated from the data by GCV or another smoothness selection criterion
- The upper bound on this estimate is , minus one due to identifiability constraint on each smooth term
s(..., bs = "cr", k = 20)Finer control of gam(): the gamma parameter
GCV is known to have some tendency to overfitting
Inside the
gam()function, the argumentgammacan increase the amount of smoothing- The default value for
gammais 1 - We can use a higher value to avoid overfitting,
gamma = 1.5, without compromising model fit
- The default value for
References
- Wood, Simon N. (2017), Generalized Additive Models: An Introduction with R. Chapman and Hall/CRC