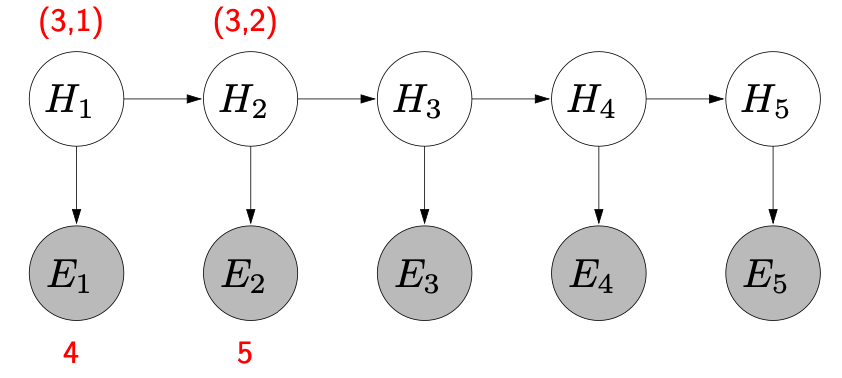
State-based vs variable-based search problems: order
There is usually an (obvious) order for state-based search problems, e.g., certain start and end points.
For variable-based search problems:
- Ordering doesn’t affect correctness (e.g., map coloring problem), so we might dynamically choose a better ordering of the variables (e.g., lookahead).
- Variables are interdependent in a local way.
Equivalent terminologies
- Variable-based models graphical models
- Probablistic graphical models = Markov networks, Bayesian networks
- Markov networks undirected graphical models
- Bayesian networks directed graphical models
Constraint Satisfaction Problems (CSPs)
Factor Graphs
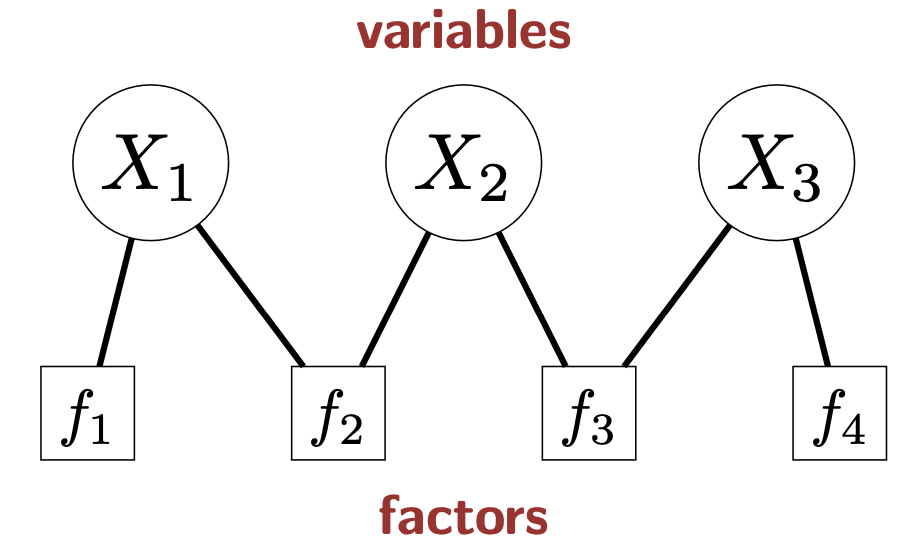
Figure source: Stanford CS221 spring 2025, lecture slides week 6
Variables (and their domains):
Factors: constraints or preferrence
- Factors measure how good an assignment is. We prefer that can achieve higher value of .
- If , it describes a constraint.
- For example, a factor for force and to be equal can be written as .
- Scope of a factor: set of variables it depends on.
- Arity of a factor: number of variables in the scope
- Unary factor: arity 1
- Binary factor: arity 2
Assignment weights: each assignment has a weight
- An assignment is consistent if Weight
Goal: find the best assignment of values to the variables to maximize the weight
- A CSP is satisfiable if .
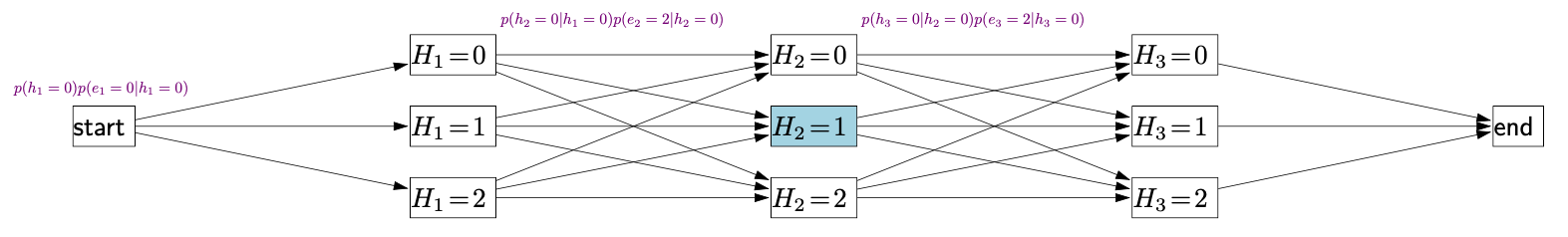
Exact Backtracking Search
- In backtracking search
- Each node is a partial assignment, and each child node is an extension of the partial assignment.
- Each leaf node is a complete assignment.
- For a partial assignment and a new variable that’s not in , dependent factor is the set of factors that only depend on and , but not on any other variables.
Backtracking search algorithm for CSPs
Backtrack(, , Domains):
- If is complete assignment: update best and return
- Choose unassigned variable (MCV)
- Order values in Domain of chosen variable (LCV)
- For each value in that order:
- If : continue (new partial assignment is inconsistent)
- Domains Domains via lookahead
- If any Domains is empty: continue
- Backtrack(, , Domains)
In the above algorithm, the blue contents are the ones that can be optimized
One-step lookahead: forward checking. After assigning a variable , eliminate inconsistent values from the dominas of ’s neighbors
Dynamic ordering
- Choose an unassigned variable: choose the most constrained variable (MCV), i.e., the variable that has the smallest domain).
- If going to fail, fail early (more pruning)
- Because we need to find an assignment for every variable
- This is useful when some factor are constraints (can prune assginemnts with weight 0)
- Order values of a selected variable: least constrained value (LCV), descending order of the sum of consistent values of neighboring variables
- Choose value that is most likely to lead to solution
- Because for each variable only need to choose some values
- Useful when all factors are constraints (Only need to find an assignment with weight 1)
Arc consistency
A variable is arc consistent wrt if for each , there exists such that for all factors whose scope contains and .
AC-3 algorithm: repeatedly enforce arc consistency on all variables
Approximate Search
- Backtracking and beam search: extend partial assignments
- Local search: modify complete assignments
Beam search
- Greedy search: we assume we have a fixed ordering of the variables. Then in every step of assigning a value to a variable, greedy search is to use the assignment with the highest weight
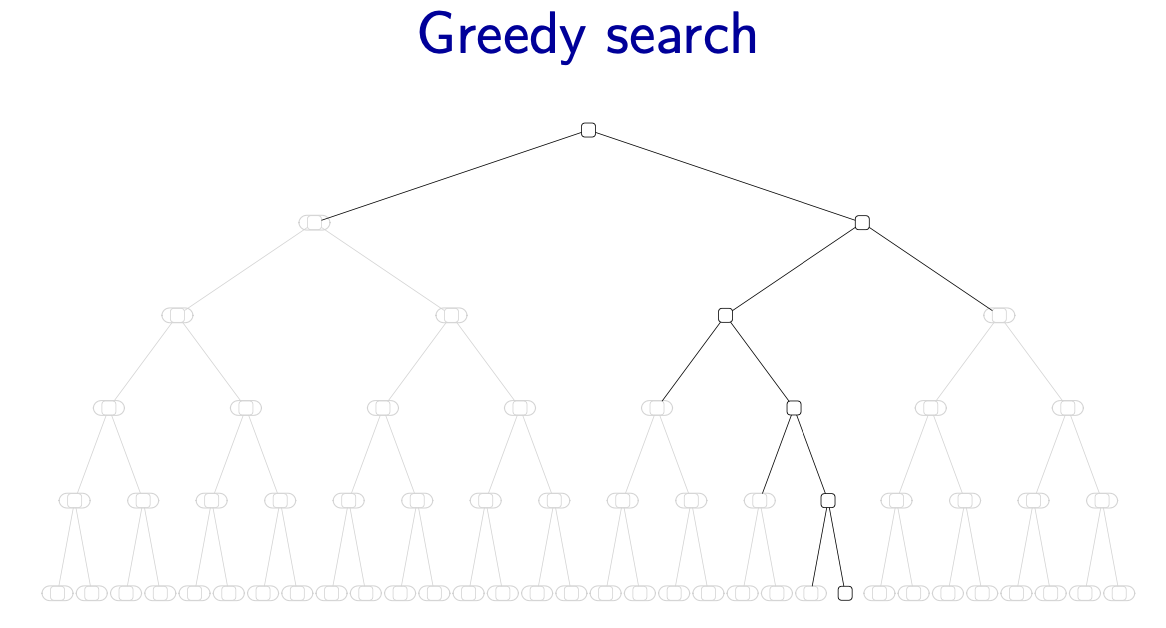
Figure source: Stanford CS221 spring 2025, lecture slides week 6
- Beam search: keep track of (at most) best partial assignments at each level of the search tree
- Note: these candidates are not guaranteed to be the best at each level
- Time complexity of beam search is
- Depth: number of variables
- Branching factor
- Beam size
- Beam size controls trade-off between efficiency and accuracy
- : greedy search, time
- : BFS, time
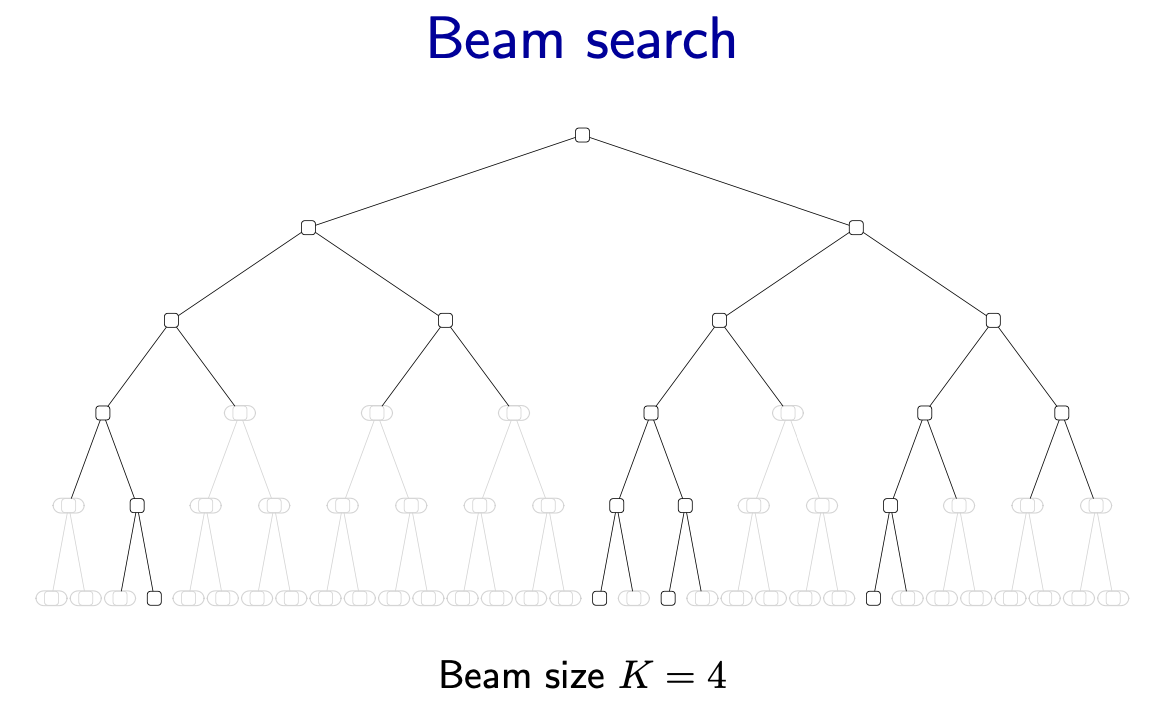
Figure source: Stanford CS221 spring 2025, lecture slides week 6
Local search
The goal is to improve an old complete assignment.
Locality: When evaluating possible re-assignments to , only need to consider the factors that depend on .
Iterated conditional modes (ICM) algorithm
- Initialize to a random complete assignment
- Loop through until convergence:
- Compute weight of for each
- with highest weights
ICM can stuck at local optima
ICM has linear time complexity
Markov Networks
A Markov network is a factor graph which defines a joint distribution over random variables : where is the normalization constant.
- Markov network factor graphs probability
Marginal probability of is
Gibbs Sampling
- Gibbs sampling algorithm:
- Initialize to a random complete assignment
- Loop through until convergence:
- Set with probability
- Increment count
- Estimate
- Search vs sampling
| Iterated Conditional Modes | Gibbs Sampling |
|---|---|
| maximum weight assignment | marginal probabilities |
| choose best value | sample a value |
| converges to local optimum | marginals converge to correct answer |
Bayesian Networks
- Markov networks vs Bayesian networks
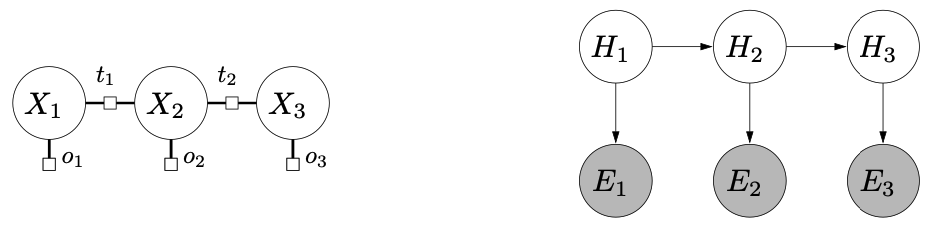
Figure source: Stanford CS221 spring 2025, lecture slides week 7
| Markov networks | Bayesian networks |
|---|---|
| arbitrary factors | local conditional probabilities |
| set of preferences | generative process |
| un-directed graphs | directed graphs |
Let be random variables. A Bayesian network is a directed acyclic graph (DAG) that specifies a joint distribution over as a project of local conditional distributions, one for each node
Reducing Bayesian networks to Markov networks
- Remember to have a single factor connecting each parent.
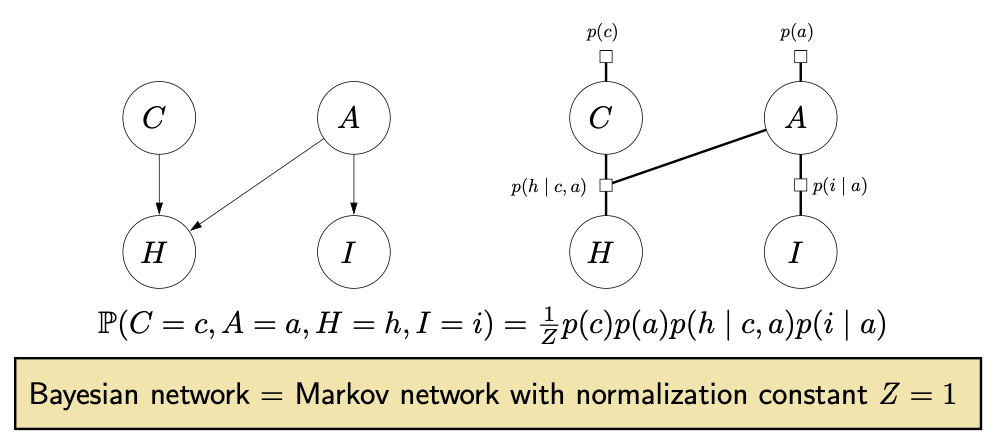
Figure source: Stanford CS221 spring 2025, lecture slides week 7
- Leveraging additional structure
- Throw away any unobserved leaves before running inference
- Throw away any disconnected components before running inference (independence)
Parameter Estimation
Smoothing
- Laplace smoothing: for each distribution and partial assignment , add to count
- This is like adding a Dirichlet prior
Expectation Maximization (EM) Algorithm
- Initialize randomly
- Repeat until converge
- E step: fix , update
- For each compute
- Create fully-observed weighted examples: with weight
- M step: fix , update
- Maximum likelihood (count and normalize) on weighted examples to get
- E step: fix , update
- Properties of the EM algorithm
- EM algorithm deals with hidden variables
- Intuition: generalization of the K-means algorithm:
- Cluster centroids = parameter
- Cluster assignments = hidden variables
- EM algorithm converges to local optima
A more general version of the EM algorithm
Choose the initial parameters
E step: since the conditional posterior contains all of our knowledge about the latent variable , we compute the expected complete-data log likelihood under it.
M step: revise parameter estimate
- Note in the maximizing step, the logarithm acts driectly on the joint likelihood , so the maximizating will be tractable.
Check for convergence of the log likelihood or the parameter values. If not converged, use to replace , and return to step 2.
See my EM algorithm notes for more details